Making Mosaics of SOXS Images¶
Warning
This is a currently experimental feature–please report any bugs to the SOXS issues page.
Note
This functionality requires the reproject package to be installed.
SOXS has two functions to create a mosaic of simulated X-ray observations from
the same source. To create a mosaic event files from a single source, use the
make_mosaic_events() function. We’ll use a SIMPUT catalog
from a Chandra image of Cas A (which you can get
here,
and make four pointings using the Lynx LXM.
import soxs
# This is a list of tuples of RA, Dec aimpoints of the different
# pointings
pointing_list = [
(350.917572, 58.79338837),
(350.755960, 58.79442533),
(350.914650, 58.87563736),
(350.755584, 58.87498051)
]
obs_list = soxs.make_mosaic_events(pointing_list, "casa.simput", "casa",
(1.0,"ks"), "lynx_lxm", overwrite=True)
make_mosaic_events() takes nearly all of the parameters
which can be supplied to the instrument_simulator()
function, see Mosaic API for details.
For the list of pointings, one could also create an ASCII table that contains
the list of pointings that looks like this, and supply the filename as the
pointing_list argument:
# RA Dec
350.917572 58.79338837
350.755960 58.79442533
350.914650 58.87563736
350.755584 58.87498051
What is returned in obs_list is an ASCII table of RA, Dec and the name of
the event file corresponding to each pointing. To create a single mosaic image
from these event files, this list needs to be fed into the
make_mosaic_image() function:
soxs.make_mosaic_image(obs_list, "casa_all_image.fits", overwrite=True)
Which will create an image of each pointing, save it to disk, and then combine them into a final image.
Optionally, if you provide a filename to the evt_file argument, the events from
the separate files will be combined into a single event file:
soxs.make_mosaic_image(obs_list, "casa_all_image.fits", evt_file="casa_all_evt.fits",
overwrite=True)
make_mosaic_image() takes many parameters which can be
supplied to write_image(), and also allows one to create an
exposure map, so it also takes several parameters which are supplied to
make_exposure_map(). Here is an example of making a mosaicked
image which has been reblocked by a factor of 2 and created with an exposure map:
soxs.make_mosaic_image(obs_list, "casa_all_image.fits", overwrite=True,
use_expmap=True, expmap_energy=1.5, reblock=2)
In this case, an exposure map is made for each pointing, these files are combined into a single exposure map, and the mosaicked counts image is divided by the map to create a flux image, which is also saved to disk. These are the results in ds9:
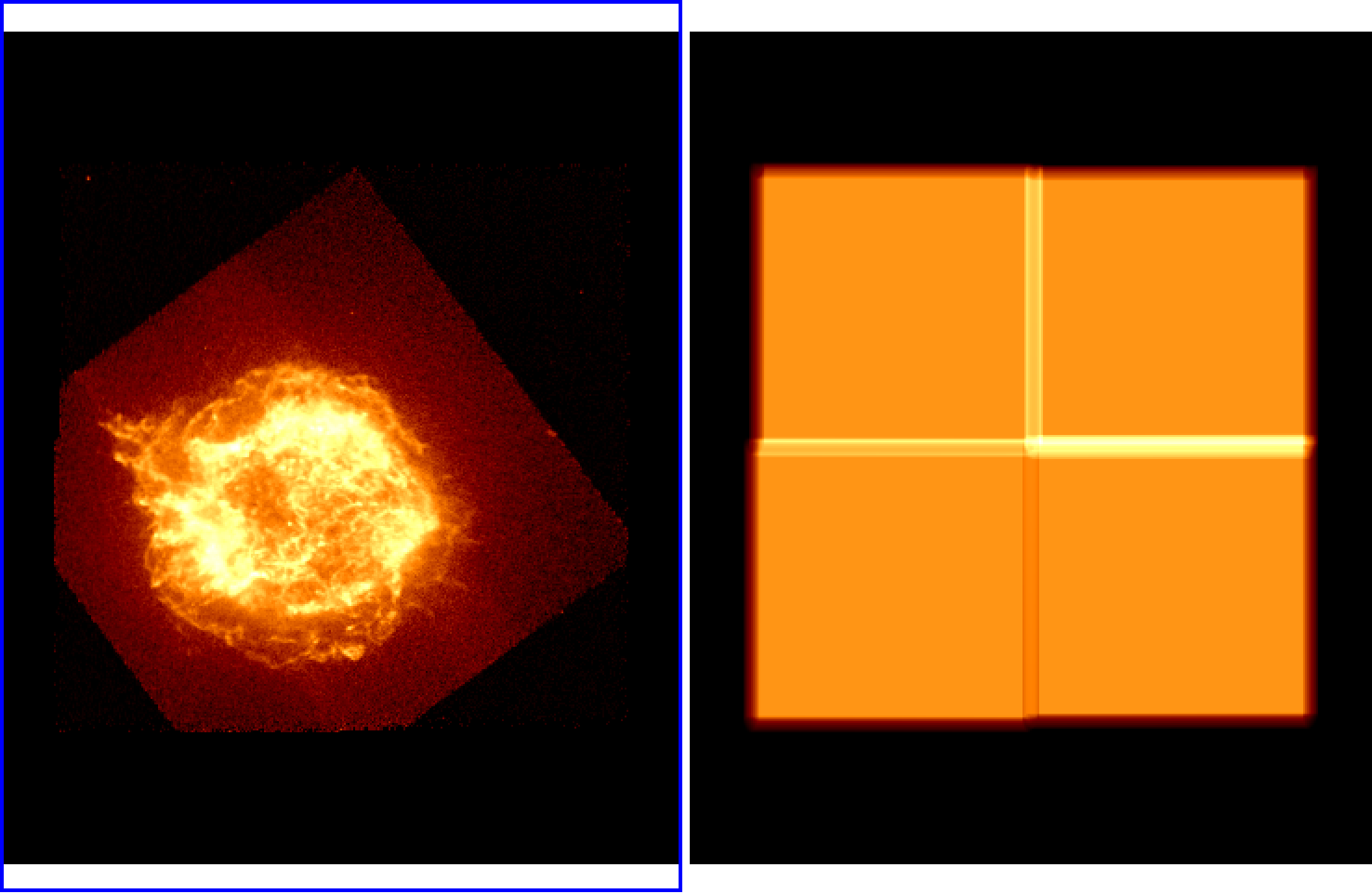
For other options, see Mosaic API.