Last modified: February 26, 2007
This thread uses
PINTofALE
to generate Suzaku/XIS spectra and Suzaku/HXD count rate estimates
for a coronal source, using a specified multi-temperature DEM,
normalized to a Chandra/ACIS-I count rate.
The Suzaku ARF, RMF, and background files used here can be obtained from
heasarc.gsfc.nasa.gov/docs/astroe/prop_tools/xis_mat.html
for XIS and
heasarc.gsfc.nasa.gov/docs/astroe/prop_tools/hxd_mat.html
for HXD.
All the steps described here are repeated in the .par file: Suzaku_ao2.par.
# start IDL idl ; set up the PoA environment ; .run initale ;NOTE: ; if INITALE fails, run the script ; @PoA_constructor
Set up the local environment variables, such as pathnames specific to the installation, source model parameters, and observation specific variables.
pimmsdir = '/soft/pimms/data' ; change to your local installation
!LDBDIR = '$SPEX' ; Atomic Line Database
; choose from the predefined '$CHIANTI', '$SPEX', '$APED',
; or specify the full path name to the line database
!IONEQF = 'ioneq/mazzotta_etal.ioneq' ; ion balance
!CEROOT = 'chianti' ; 'cie' or 'chianti'
!NH = 3e20 ; H column density [cm^-2]
!EDENS = 1.0e9 ; electron number density [cm^-3]
!ABUND = getabund('asplund et al.') ; element abundances
T_components = [6.1, 6.8, 7.2] ; log(T[K]) components in EM
EM_components = [6.1d11, 6.1d11, 7.1e11] ; Emission Measure [cm^-5]
EXPTIME = 30. ; nominal exposure time [ks]
acisi_rate = 0.1 ; Chandra/ACIS-I rate [ct/s] - 0 or less to ignore
acisi_emin = 0.3 ; minimum ACIS-I energy to consider [keV]
acisi_emax = 8.0 ; maximum ACIS-I energy to consider [keV]
cornorm = 0.0 ; background correction norm
Provide the names of the files containing the ARFs, RMFs, and
background PHAs. Note that 'none' is an acceptable option.
XIS related files can be obtained from
heasarc.gsfc.nasa.gov/docs/astroe/prop_tools/xis_mat.html
and HXD related files from
heasarc.gsfc.nasa.gov/docs/astroe/prop_tools/hxd_mat.html.
xisFI_RMF = 'ae_xi2_20051219.rmf' ; XIS FI RMF filename xisFI_ARF = 'ae_xi2_onaxis_20050916.arf' ; XIS FI ARF filename xisFI_BKG = 'ae_xi2_back.pha' ; XIS FI bkg PHA filename ; xisBI_RMF = 'ae_xi1_20051210d.rmf' ; XIS BI RMF filename xisBI_ARF = 'ae_xi1_onaxis_20050916.arf' ; XIS BI ARF filename xisBI_BKG = 'ae_xi1_back.pha' ; XIS BI bkg PHA filename hxdPIN_RSP = 'ae_hxd_pinxinom_20051104.rsp' ;HXD PIN RSP filename hxdPIN_BKG = 'ae_hxd_pinbkg_20051105.pha' ;HXD PIN bkg PHA filename ; hxdGSO_RSP = 'ae_hxd_gso_20051019.rsp' ;HXD GSO RSP filename hxdGSO_BKG = 'ae_hxd_gsobkg_20051105.pha' ;HXD GSO bkg PHA filename
These variables will be computed or determined downstream, but this thread is set up to speed up calculations by not repeating unnecessary tasks. Unless explicitly uncommented, inputs corresponding to the following will not be read in again after the first run.
; ae2_lstr = 0 ;the line emissivity database structure ; ae2_cstr = 0 ;the XIS continuum emissivity database structure ; ae2_hcstr = 0 ;the HXD continuum emissivity database structure ; rm_xisFI = 0 ;the XIS/FI response matrix ; ea_xisFI = 0 ;the XIS/FI effective area ; bg_xisFI = 0 ;the XIS/FI background spectrum ; rm_xisBI = 0 ;the XIS/BI response matrix ; ea_xisBI = 0 ;the XIS/BI effective area ; bg_xisBI = 0 ;the XIS/BI background spectrum ; rm_hxdPIN = 0 ;the HXD/PIN response matrix ; bg_hxdPIN = 0 ;the HXD/PIN background spectrum ; rm_hxdGSO = 0 ;the HXD/GSO response matrix ; bg_hxdGSO = 0 ;the HXD/GSO background spectrum
A Differential Emission Measure (DEM) is required to estimate the amount of emission at various temperatures. Typically, a 2-temperature model is used. Here we will use PINTofALE's mk_dem(), which constructs a DEM array given a temperature grid and emission measure components. We use as the temperature grid !LOGT. The emission measure components are T_components and EM_components as defined above
!DEM=mk_dem('delta', logT = !LOGT, pardem=T_components, indem=EM_components)
; XIS/FI
dwvl=0.000434 & xis_FIwgrid=10.^(findgen(4097)*dwvl)
if strlowcase(xisFI_RMF) ne 'none' and n_tags(rm_xisFI) eq 0 then $
rm_xisFI=rd_ogip_rmf(xisFI_RMF,effar=qe_xisFI)
if n_tags(rm_xisFI) ne 0 then xis_FIwgrid=12.3985/[min(rm_xisFI.ELO),rm_xisFI.EHI]
if strlowcase(xisFI_RMF) eq 'none' and n_tags(rm_xisFI) eq 0 then qe_xisFI=0.*xis_FIwgrid[1:*]+1.
if strlowcase(xisFI_ARF) ne 'none' and not keyword_set(ea_xisFI) then ea_xisFI=rdarf(xisFI_ARF,ea_xisFIstr)
if strlowcase(xisFI_ARF) eq 'none' and not keyword_set(ea_xisFI) then ea_xisFI=0.*xis_FIwgrid[1:*]+1.
if strlowcase(xisFI_BKG) ne 'none' and n_tags(bg_xisFI) eq 0 then bg_xisFI=mrdfits(xisFI_BKG,1,hbg_xisFI)
if strlowcase(xisFI_BKG) eq 'none' and n_tags(bg_xisFI) eq 0 then $
bg_xisFI=create_struct('CHANNEL',lindgen(n_elements(xis_FIwgrid)-1),'COUNTS',lonarr(0*xis_FIwgrid[1:*]))
bg_xisFI_exp=1. & if keyword_set(hbg_xisFI) then bg_xisFI_exp=float(sxpar(hbg_xisFI,'EXPOSURE'))>1.
; XIS/BI
dwvl=0.000434 & xis_BIwgrid=10.^(findgen(4097)*dwvl)
if strlowcase(xisBI_RMF) ne 'none' and n_tags(rm_xisBI) eq 0 then $
rm_xisBI=rd_ogip_rmf(xisBI_RMF,effar=qe_xisBI)
if n_tags(rm_xisBI) ne 0 then xis_BIwgrid=12.3985/[min(rm_xisBI.ELO),rm_xisBI.EHI]
if strlowcase(xisBI_RMF) eq 'none' and n_tags(rm_xisBI) eq 0 then qe_xisBI=0.*xis_BIwgrid[1:*]+1.
if strlowcase(xisBI_ARF) ne 'none' and not keyword_set(ea_xisBI) then ea_xisBI=rdarf(xisBI_ARF,ea_xisBIstr)
if strlowcase(xisBI_ARF) eq 'none' and not keyword_set(ea_xisBI) then ea_xisBI=0.*xis_BIwgrid[1:*]+1.
if strlowcase(xisBI_BKG) ne 'none' and n_tags(bg_xisBI) eq 0 then bg_xisBI=mrdfits(xisBI_BKG,1,hbg_xisBI)
if strlowcase(xisBI_BKG) eq 'none' and n_tags(bg_xisBI) eq 0 then $
bg_xisBI=create_struct('CHANNEL',lindgen(n_elements(xis_BIwgrid)-1),'COUNTS',lonarr(0*xis_BIwgrid[1:*]))
bg_xisBI_exp=1. & if keyword_set(hbg_xisBI) then bg_xisBI_exp=float(sxpar(hbg_xisBI,'EXPOSURE'))>1.
; HXD/PIN
dwvl=0.0094 & hxd_PINwgrid=10.^(findgen(257)*dwvl-0.89)
if strlowcase(hxdPIN_RSP) ne 'none' and n_tags(rm_hxdPIN) eq 0 then $
rm_hxdPIN=rd_ogip_rmf(hxdPIN_RSP,effar=ea_hxdPIN)
if n_tags(rm_hxdPIN) ne 0 then hxd_PINwgrid=12.3985/[min(rm_hxdPIN.ELO),rm_hxdPIN.EHI]
if n_tags(rm_hxdPIN) ne 0 then ea_hxdPIN=total(rm_hxdPIN.MATRIX,1) else ea_hxdPIN=0.*hxd_PINwgrid[1:*]+1.
if strlowcase(hxdPIN_BKG) ne 'none' and n_tags(bg_hxdPIN) eq 0 then bg_hxdPIN=mrdfits(hxdPIN_BKG,1,hbg_hxdPIN)
if strlowcase(hxdPIN_BKG) eq 'none' and n_tags(bg_hxdPIN) eq 0 then $
bg_hxdPIN=create_struct('CHANNEL',lindgen(n_elements(hxd_PINwgrid)-1),'COUNTS',lonarr(0*hxd_PINwgrid[1:*]))
bg_hxdPIN_exp=1. & if keyword_set(hbg_hxdPIN) then bg_hxdPIN_exp=float(sxpar(hbg_hxdPIN,'EXPOSURE'))>1.
; HXD/GSO
dwvl=0.0059 & hxd_GSOwgrid=10.^(findgen(513)*dwvl-1.92)
if strlowcase(hxdGSO_RSP) ne 'none' and n_tags(rm_hxdGSO) eq 0 then $
rm_hxdGSO=rd_ogip_rmf(hxdGSO_RSP,effar=ea_hxdGSO)
if n_tags(rm_hxdGSO) ne 0 then hxd_GSOwgrid=12.3985/[min(rm_hxdGSO.ELO),rm_hxdGSO.EHI]
if n_tags(rm_hxdGSO) ne 0 then ea_hxdGSO=total(rm_hxdGSO.MATRIX,1) else ea_hxdGSO=0.*hxd_GSOwgrid[1:*]+1.
if strlowcase(hxdGSO_BKG) ne 'none' and n_tags(bg_hxdGSO) eq 0 then bg_hxdGSO=mrdfits(hxdGSO_BKG,1,hbg_hxdGSO)
if strlowcase(hxdGSO_BKG) eq 'none' and n_tags(bg_hxdGSO) eq 0 then $
bg_hxdGSO=create_struct('CHANNEL',lindgen(n_elements(hxd_GSOwgrid)-1),'COUNTS',lonarr(0*hxd_GSOwgrid[1:*]))
bg_hxdGSO_exp=1. & if keyword_set(hbg_hxdGSO) then bg_hxdGSO_exp=float(sxpar(hbg_hxdGSO,'EXPOSURE'))>1.
wmin=min(xis_BIwgrid) < min(xis_FIwgrid)
wmax=max(xis_BIwgrid) < max(xis_FIwgrid)
incieq=1
if strpos(strlowcase(!LDBDIR),'apec',0) ge 0 then incieq=0 else $
if strpos(strlowcase(!LDBDIR),'aped',0) ge 0 then incieq=0 else $
if strpos(strlowcase(!LDBDIR),'gu',0) ge 0 then incieq=0
if n_tags(ae2_lstr) eq 0 then $
ae2_lstr=rd_list('all|'+strtrim(wmin,2)+'-'+strtrim(wmax,2)+'|'+!LDBDIR,sep='|',$
incieq=incieq,n_e=!EDENS,eqfile=!IONEQF,verbose=!VERBOSE)
; If !LDBDIR is set to APED, Anders & Grevesse abundances
; are already included in the emissivities. In which case,
; redefine the abundance array to be relative to APED.
abnorm=0*!ABUND+1.
if strpos(strlowcase(!LDBDIR),'apec',0) ge 0 or $
strpos(strlowcase(!LDBDIR),'aped',0) ge 0 then $
abnorm=getabund('anders & grevesse')
if n_tags(ae2_cstr) eq 0 then $
fc=rd_cont(!CEROOT,n_e=!EDENS,wrange=[wmin,wmax],dbdir=!CDBDIR,$
abund=!ABUND,fcstr=ae2_cstr,twoph=twoph)
if n_tags(ae2_hcstr) eq 0 then $
fc=rd_cont(!CEROOT,n_e=!EDENS,wrange=12.3985/[60.,1.],dbdir=!CDBDIR,$
abund=!ABUND,fcstr=ae2_hcstr,twoph=twoph)
; line intensities linint= lineflx(ae2_lstr.LINE_INT,ae2_lstr.LOGT,abs(ae2_lstr.WVL),$ ae2_lstr.Z,DEM=!DEM,abund=!ABUND/abnorm) ;[ph/s/cm^2] ; continuum intensities conint= lineflx(ae2_cstr.CONT_INT,ae2_cstr.LOGT,abs(ae2_cstr.midWVL),$ DEM=!DEM) ;[ph/s/cm^2/Ang] cwb= mid2bound(ae2_cstr.midWVL) cdw= cwb[1:*]-cwb conint= conint*cdw ;[ph/s/cm^2] ; continuum intensities for HXD hconint=lineflx(ae2_hcstr.CONT_INT,ae2_hcstr.LOGT,abs(ae2_hcstr.midWVL),$ DEM=!DEM) ;[ph/s/cm^2/Ang] cwb= mid2bound(ae2_hcstr.midWVL) cdw= cwb[1:*]-cwb hconint=hconint*cdw ;[ph/s/cm^2] ; ISM absorption ltau= ismtau(abs(ae2_lstr.WVL),NH=!NH,fH2=!fH2,He1=!He1,HeII=!HeII,$ Fano=Fano,wam=wam,/bam,abund=!ABUND,verbose=!VERBOSE) ctau= ismtau(abs(ae2_cstr.midWVL),NH=!NH,fH2=!fH2,He1=!He1,HeII=!HeII,$ Fano=Fano,wam=wam,/bam,abund=!ABUND,verbose=!VERBOSE) hctau= ismtau(abs(ae2_hcstr.midWVL),NH=!NH,fH2=!fH2,He1=!He1,HeII=!HeII,$ Fano=Fano,wam=wam,/bam,abund=!ABUND,verbose=!VERBOSE) ltrans= exp(-ltau) ctrans= exp(-ctau) hctrans=exp(-hctau) ; line fluxes linflx= linint * ltrans ; [ph/s/cm^2] conflx= conint * ctrans ; [ph/s/cm^2] hconflx=hconint * hctrans ; [ph/s/cm^2]
We will assume that a Chandra/ACIS-I count rate is available, and require that the simulation match this rate. Data from other missions such as ROSAT, ASCA, BeppoSAX, etc. can be dealt with in the same manner.
; First find and read in the Chandra/ACIS-I effective area
; you will need to know where your local PIMMS installation
; is to do this.
rd_pimms_file,get_pimms_file('chandra','acis-i',pdir=pimmsdir),$
acisi_effar,acisi_wvlar,/wave
; fold in effective area
acisi_wmin=12.3985/acisi_emax & acisi_wmax=12.3985/acisi_emin
os=sort(acisi_wvlar) & acisi_wvlar=acisi_wvlar[os] & acisi_effar=acisi_effar[os]
;
lwvl=abs(ae2_lstr.WVL) & ow=where(lwvl ge acisi_wmin and lwvl le acisi_wmax)
effar=(interpol(acisi_effar,acisi_wvlar,lwvl[ow])>0) < max(acisi_effar)
flxlin= linflx[ow]*effar ;[ct/s]
;
cwvl=abs(ae2_cstr.midWVL) & ow=where(lwvl ge acisi_wmin and lwvl le acisi_wmax)
effar=(interpol(acisi_effar,acisi_wvlar,cwvl)>0) < max(acisi_effar)
flxcon= conflx[ow]*effar ;[ct/s]
; total predicted rate in Chandra/ACIS-I
pred_rate=total(flxlin)+total(flxcon)
print,''
if acisi_rate gt 0 then $
print,'Rescaling input DEM by a factor '+strtrim(acisi_rate/pred_rate,2)
print,'to match required ACIS-I rate of '+strtrim(acisi_rate,2)+' ct/s'
print,''
rescale_factor=1.0
if acisi_rate gt 0 then rescale_factor = acisi_rate/pred_rate
!DEM = !DEM * rescale_factor
linflx = linflx * rescale_factor
conflx = conflx * rescale_factor
hconflx = hconflx * rescale_factor
; HXD/PIN wv_hxdPIN=0.5*(hxd_PINwgrid[1:*]+hxd_PINwgrid) ; lwvl=abs(ae2_lstr.WVL) effar=(interpol(ea_hxdPIN,wv_hxdPIN,lwvl)>0) < max(ea_hxdPIN) flxlin= linflx*effar ;[ct/s] ; cwvl=abs(ae2_hcstr.midWVL) effar=(interpol(ea_hxdPIN,wv_hxdPIN,cwvl)>0) < max(ea_hxdPIN) flxcon= hconflx*effar ;[ct/s] ; flx_hxdPIN = total(flxlin)+total(flxcon) ; HXD/GSO wv_hxdGSO=0.5*(hxd_GSOwgrid[1:*]+hxd_GSOwgrid) ; lwvl=abs(ae2_lstr.WVL) effar=(interpol(ea_hxdGSO,wv_hxdGSO,lwvl)>0) < max(ea_hxdGSO) flxlin= linflx*effar ;[ct/s] ; cwvl=abs(ae2_hcstr.midWVL) effar=(interpol(ea_hxdGSO,wv_hxdGSO,cwvl)>0) < max(ea_hxdGSO) flxcon= hconflx*effar ;[ct/s] ; flx_hxdGSO = total(flxlin)+total(flxcon) print,'' print,'HXD/PIN bremss count rate = ',flx_hxdPIN,' total counts = ',flx_hxdPIN*EXPTIME*1e3 print,'HXD/PIN background counts = ',(1.+cornorm)*total(bg_HXDPIN.COUNTS)*EXPTIME*1e3/bg_hxdPIN_exp print,'' print,'HXD/GSO bremss count rate = ',flx_hxdGSO,' total counts = ',flx_hxdGSO*EXPTIME*1e3 print,'HXD/GSO background counts = ',(1.+cornorm)*total(bg_HXDGSO.COUNTS)*EXPTIME*1e3/bg_hxdGSO_exp print,''
; XIS/FI wv_XISFI=0.5*(XIS_FIwgrid[1:*]+XIS_FIwgrid) ; lwvl=abs(ae2_lstr.WVL) effar=(interpol(ea_XISFI,wv_XISFI,lwvl)>0) < max(ea_XISFI) flxlin= linflx*effar ;[ct/s] ; cwvl=abs(ae2_hcstr.midWVL) effar=(interpol(ea_XISFI,wv_XISFI,cwvl)>0) < max(ea_XISFI) flxcon= hconflx*effar ;[ct/s] ; flx_XISFI = total(flxlin)+total(flxcon) ; XIS/BI wv_XISBI=0.5*(XIS_BIwgrid[1:*]+XIS_BIwgrid) ; lwvl=abs(ae2_lstr.WVL) effar=(interpol(ea_XISBI,wv_XISBI,lwvl)>0) < max(ea_XISBI) flxlin= linflx*effar ;[ct/s] ; cwvl=abs(ae2_hcstr.midWVL) effar=(interpol(ea_XISBI,wv_XISBI,cwvl)>0) < max(ea_XISBI) flxcon= hconflx*effar ;[ct/s] ; flx_XISBI = total(flxlin)+total(flxcon) print,'' print,'XIS/FI count rate = ',flx_XISFI,' total counts = ',flx_XISFI*EXPTIME*1e3 print,'XIS/FI background counts = ',(1.+cornorm)*total(bg_XISFI.COUNTS)*EXPTIME*1e3/bg_hxdPIN_exp print,'' print,'XIS/BI count rate = ',flx_XISBI,' total counts = ',flx_XISBI*EXPTIME*1e3 print,'XIS/BI background counts = ',(1.+cornorm)*total(bg_XISBI.COUNTS)*EXPTIME*1e3/bg_hxdPIN_exp print,''
To construct the Suzaku/XIS spectra, we use the line and continuum fluxes calculated above (LINFLX and CONFLX), rebin them into a consistent energy grid, and convolve with the response matrix.
; XIS/FI ; bin the spectra egrid=12.3985/xis_FIwgrid & os=sort(egrid) & egrid=egrid[os] & tmp=egrid lnrg=12.3985/abs(ae2_lstr.WVL) & os=sort(lnrg) xisFI_lspc= hastogram(lnrg[os],egrid,wts=linflx[os]) ;[ph/s/cm^2/bin] ; egrid=12.3985/xis_FIwgrid & os=sort(egrid) & egrid=egrid[os] cnrg=12.3985/ae2_cstr.WVL & os=sort(cnrg) xisFI_cspc= rebinw(conflx[os],cnrg[os],egrid,/perbin) ;[ph/s/cm^2/bin] ; xisFI_spc= xisFI_lspc + xisFI_cspc ; convolve with RMF tmp=12.3985/xis_FIwgrid & ee_xisFI=0.5*(tmp[1:*]+tmp) if strlowcase(xisFI_RMF) ne 'none' then $ conv_rmf,egrid,xisFI_spc,xisFI_channel,xisFI_predct,rm_xisFI,$ effar=ea_xisFI,nrgar=0.5*(ea_xisFIstr.ELO+ea_xisFIstr.EHI),$ verbose=!VERBOSE ; units are now in ct/s/bin, so multiply by EXPTIME xisFI_predct = xisFI_predct * EXPTIME * 1e3 ;[ct/bin] ; XIS/BI ; bin the spectra egrid=12.3985/xis_BIwgrid & os=sort(egrid) & egrid=egrid[os] & tmp=egrid lnrg=12.3985/abs(ae2_lstr.WVL) & os=sort(lnrg) xisBI_lspc= hastogram(lnrg[os],egrid,wts=linflx[os]) ;[ph/s/cm^2/bin] ; egrid=12.3985/xis_BIwgrid & os=sort(egrid) & egrid=egrid[os] cnrg=12.3985/ae2_cstr.WVL & os=sort(cnrg) xisBI_cspc= rebinw(conflx[os],cnrg[os],egrid,/perbin) ;[ph/s/cm^2/bin] ; xisBI_spc= xisBI_lspc + xisBI_cspc ; convolve with RMF tmp=12.3985/xis_BIwgrid & ee_xisBI=0.5*(tmp[1:*]+tmp) if strlowcase(xisBI_RMF) ne 'none' then $ conv_rmf,egrid,xisBI_spc,xisBI_channel,xisBI_predct,rm_xisBI,$ effar=ea_xisBI,nrgar=0.5*(ea_xisBIstr.ELO+ea_xisBIstr.EHI),$ verbose=!VERBOSE ; units are now in ct/s/bin, so multiply by EXPTIME xisBI_predct = xisBI_predct * EXPTIME * 1e3 ;[ct/bin]
nFI=n_elements(xisFI_predct) xisFI_counts=lonarr(nFI) & xisFI_countsbg=lonarr(nFI) & xisFI_countstot=lonarr(nFI) tmp=xisFI_predct & for i=0L,nFI-1L do if tmp[i] gt 1d-20 then $ xisFI_counts[i]=randomu(seed,poisson=tmp[i]) ; tmp=bg_xisFI.counts*(EXPTIME*1e3)*(1.+cornorm)/BG_XISFI_EXP for i=0L,nFI-1L do if tmp[i] gt 1d-20 then $ xisFI_countsbg[i]=randomu(seed,poisson=tmp[i]) ; xisFI_countstot = xisFI_counts + xisFI_countsbg nBI=n_elements(xisBI_predct) xisBI_counts=lonarr(nBI) & xisBI_countsbg=lonarr(nBI) & xisBI_countstot=lonarr(nBI) tmp=xisBI_predct & for i=0L,nBI-1L do if tmp[i] gt 1d-20 then $ xisBI_counts[i]=randomu(seed,poisson=tmp[i]) ; tmp=bg_xisBI.counts*(EXPTIME*1e3)*(1.+cornorm)/BG_XISBI_EXP for i=0L,nBI-1L do if tmp[i] gt 1d-20 then $ xisBI_countsbg[i]=randomu(seed,poisson=tmp[i]) ; xisBI_countstot = xisBI_counts + xisBI_countsbg
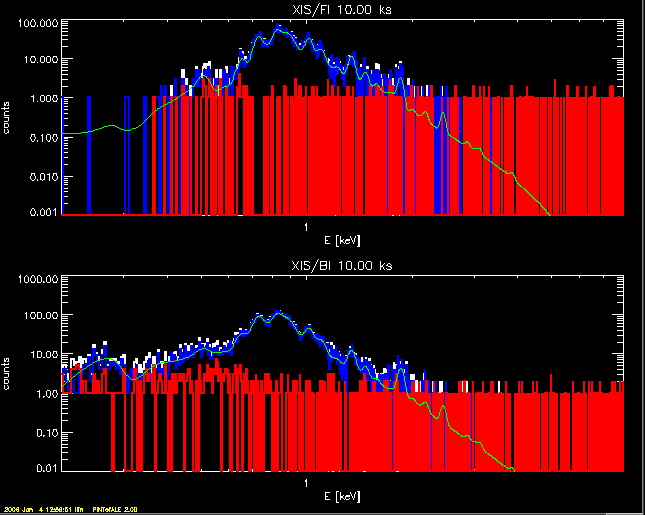
peasecolr & !p.multi=[0,1,2] plot,xisFI_channel,xisFI_countstot,psym=10,thick=2,$ xtitle='E [keV]',ytitle='counts',$ title='XIS/FI '+strtrim(string(EXPTIME,'(f5.2)'),2)+' ks',$ xrange=[0.2,8],/xs,/xlog,yrange=max(xisFI_countstot)*[1e-4,1],/ylog oplot,xisFI_channel,xisFI_counts,psym=10,thick=2,col=1 oplot,xisFI_channel,xisFI_countsbg,psym=10,thick=2,col=2 oplot,xisFI_channel,xisFI_predct,col=3 plot,xisBI_channel,xisBI_countstot,psym=10,thick=2,$ xtitle='E [keV]',ytitle='counts',$ title='XIS/BI '+strtrim(string(EXPTIME,'(f5.2)'),2)+' ks',$ xrange=[0.2,8],/xs,/xlog,yrange=max(xisBI_countstot)*[1e-4,1],/ylog oplot,xisBI_channel,xisBI_counts,psym=10,thick=2,col=1 oplot,xisBI_channel,xisBI_countsbg,psym=10,thick=2,col=2 oplot,xisBI_channel,xisBI_predct,col=3 stample,stacol=4 !p.multi=0
------------------------------------------------------------------------------ Outputs are in the following variables: ------------------------------------------------------------------------------ ----------------------------------------------------------------------------- XIS/FI -- model spectrum, pre-RMF : xisFI_spc [ph/s/cm^2/bin] (egrid,xis_FIwgrid) total model count rate : flx_XISFI [ph/s] predicted counts intensity : xisFI_predct [ct/channel] (xisFI_channel) simulated source counts : xisFI_counts [ct/channel] (xisFI_channel) simulated background counts : xisFI_countsbg [ct/channel] (xisFI_channel) simulated total counts : xisFI_countstot [ct/channel] (xisFI_channel) ----------------------------------------------------------------------------- XIS/BI -- model spectrum, pre-RMF : xisBI_spc [ph/s/cm^2/bin] (egrid,xis_BIwgrid) total model count rate : flx_XISBI [ph/s] predicted counts intensity : xisBI_predct [ct/channel] (xisBI_channel) simulated source counts : xisBI_counts [ct/channel] (xisBI_channel) simulated background counts : xisBI_countsbg [ct/channel] (xisBI_channel) simulated total counts : xisBI_countstot [ct/channel] (xisBI_channel) ------------------------------------------------------------------------------ HXD/PIN -- bremss count rate : flx_hxdPIN [ph/s] ------------------------------------------------------------------------------ HXD/GSO -- bremss count rate : flx_hxdGSO [ph/s] ------------------------------------------------------------------------------
; Results using this thread are roughly consistent with
; PIMMS results. PIMMS returned the following XMM count rates
; for a CHANDRA-ACIS-I count rate of 1 cts/s and a Raymond-Smith
; model of temperature 0.485 keV:
; SUZAKU XIS FI : 5.066E-01 cts/s
; SUZAKU XIS BI : 1.085E+00 cts/s
;
; Running this thread with !ABUND = getabund('Allen'),
; T_components = [6.8], and obs_rate = 1.0 gives:
; SUZAKU XIS FI : 0.482 cts/s
; SUZAKU XIS BI : 0.986 cts/s